




|
Note that rff is designed to accurately compute spin-spin coupling
constants. The chemical shifts - in the examples below - are calculated with modest accuracy using existing
methods, such as GIAO at the mPW1PW91/6-311+G(d,p) level of theory. (For more information
on the state of the art in computing chemical shifts see
CHESHIRE - CHEmical SHIft REpository
maintained by Dean Tantillo).
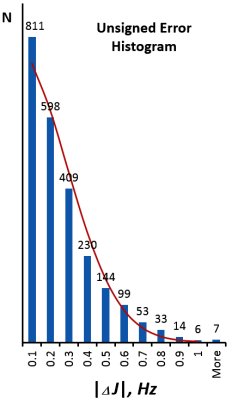
The J scaling is being refined and tested on complex organic molecules. The Unsigned Error Histrogram shows the test set as of April 2016, which contains >2400 experimental constants with rmsd = 0.28 Hz. (red line shows the ideal gaussian distribution for σ=0.28). Links to predicted NMR spectra of a few prominent natural products and other interesting molecules in the test set are found below (In parentheses after the name: rmsd(Hz) / number of reported Jexp) |
|